
Research & Initiatives
We employ unbiased proteomics approaches to understand host-pathogen interactions and map functions of understudied ubiquitin-like proteins. Many ubiquitin-like proteins are dysregulated in difficult to treat human pathologies and yet their mechanisms of action remain to be explored. Our quantitative and unbiased LC-MS/MS data has led us to studies on actin nucleation, cytokine secretion, innate immune cell function, and immunometabolism. Educating the next generation of scientists is important to us and we are open to mentoring scientists at all career stages. We look forward to exploring many new areas in cellular microbiology and immunology in the years to come.
New methods to map ubiquitin-like proteins
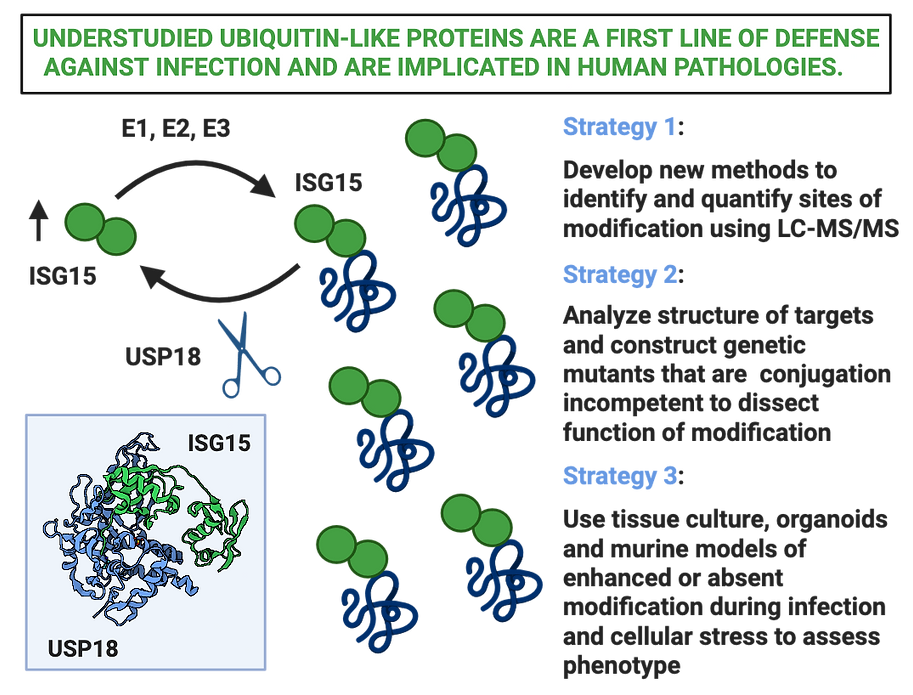
Proteoforms which encompass unique combinations of post-translationally modified proteins often act in concert to rapidly increase the functional repertoire of a cell in response to stress. Importantly, specific proteoforms contribute to and may even drive disease pathology (e.g. phosphorylated tau in Alzheimer disease). Over the past fifteen years, a sea change in sensitivity and resolution of mass spectrometry coupled with immuno-enrichment of modified protein adducts has been extremely fruitful for proteome wide, site-specific identification of acetylation, SUMOylation and ubiquitination sites using mass spectrometry. We took advantage of existing technology to enrich adducts (left by ubiquitin, ISG15 and NEDD8) in the presence or absence of ISG15 using genetically deleted mice to map the first ISGylome (Zhang et al. 2019). We are currently assessing the biological consequences of ISG15 modification following bacterial or viral infection by leveraging this resource to make ISG15 refractive targets. Going forward we are tackling site identification for other ubiquitin-like proteins as well.
.png)
How can we determine the fate of an ISGylated protein?
Ubiquitin-like modifications (UBLs) are rapid, reversible and can profoundly alter cell fate and function. Intriguingly, the majority of UBLs are involved in the cellular response to infection and are dysregulated in a number of intractable human pathologies such as cancer, autoimmune disease, protein-folding disorders and inborn errors in immunity. Upon infection, upstream immune regulators blanket host cells in Interferon Stimulated Genes (ISGs), including ISG15, however the consequences of these modifications on target protein fate and function remain unknown.
Using our method to distinguish ISG15 sites from ubiquitin sites, we identified that ISG15 modification is a much more prevalent than previously appreciated constituting up to 20% of all sites following bacterial infection in the liver (Zhang et al 2019). Since then we have compiled two additional ISGylomes following viral or bacterial infection in the lung. With this resource, we can systematically mutate lysines on ISG15 to arginines for substrates that are common to both liver and lung or unique to specific infections, in combination or one at a time. By correlating phenotypic outcomes of non-ISGylatable substrates with overarching phenotypes of enhanced or absent ISGylation, we can systematically identify the function of ISGylation during infection. Our ultimate goal is to determine, when and whether to inhibit or activate the enzymes that lead to ISGylation in difficult to treat human pathologies.
.png)
